CRISPR/Cas9 genome editing transfection reagents
OZ Biosciences offers tailored transfection solutions for CRISPR/Cas9 technology
Wide range of CRISPR knockout and activation products
Santa Cruz Biotechnology offers target-specific CRISPR/Cas9 knockout plasmids, CRISPR double nickase plasmids, CRISPR/ dCas9 activation plasmids and CRISPR lenti activation systems for over 18,910 human and 18,340 mouse protein encoding genes.
The CRISPR/Cas system is an adaptive immune defense mechanism used by Archea and bacteria for the degradation of foreign genetic material. In these organisms, the foreign genetic material from a bacteriophage is acquired and integrated into the CRISPR loci (1,2). This new material, also known as a spacer, creates a sequence-specific fragment used for future resistance against a bacteriophage infection. These sequence-specific fragments are translated into short CRISPR RNAs (crRNAs) and function as a guide to direct cleavage of complementary invading DNA via the nuclease activity of CRISPR-associated (Cas) protein also encoded by the CRISPR loci (1,2). Cas9 nuclease of the type II CRISPR system has an RNA binding domain, an alpha helix recognition lobe (REC), a nuclease lobe that include the RuvC and HNH for DNA cleavage, and a protospacer adjacent motif (PAM) interacting site (1,2). crRNA forms a complex with the Cas9 nuclease by binding to the bridge helix within the REC lobe, and forms multiple salt bridges with the backbone of the crRNA (1,2,3).
Once the crRNA binds to the Cas9 the conformation of the Cas9 nuclease changes and creates a channel that allows for DNA binding (1,2,3). The Cas9/crRNA complex scans the DNA for a PAM (5′-NGG) site (4,5,6). Recognition of a PAM site leads to unwinding of the DNA, and allows the crRNA to check for complementary DNA adjacent to the PAM site. When Cas9 binds to a PAM site adjacent to a DNA sequence that is complementary to the crRNA, the bridge helix within the REC lobe creates an RNA-DNA heteroduplex with the target DNA (3,4,7). The PAM site recognition is involved in activating the nucleolytic HNH and RuvC domains which create a double-stranded break (DSB) in the target DNA, leading to DNA degradation (1,2,5,8). If the crRNA is not complementary, then Cas9 releases and searches for another PAM site (7). Targeted genome strand breaks in the DNA can be repaired via the nonhomologous end-joining (NHEJ) repair pathway, which introduces insertion or deletions creating errors, or through the homologous directed repair (HDR) pathway, that can be used to recombine selected markers at specific sites in the genome (2,9,10). This CRISPR/Cas9 mechanism can be repurposed for genomic engineering of various systems, including mammalian cells.
Genome editing via introduction of DSBs can be performed with meganucleases, zinc finger nucleases (ZF), or transactivator-like effectors (TALEs), which recognize DNA sequences, however, each has their limitations. When using meganuclesases it is difficult to clearly show site-specific recognition between nuclease and DNA (2).The other options, ZFs and TALEs, have proven difficult to design and recognize up to 3 nt of DNA (2). Single guide RNAs (sgRNAs) that act like crRNAs are easily designed and can be expressed along with Cas9 nuclease in the same vector to target specific DNA sites for genome editing. The CRISPR/Cas 9 system also has higher sensitivity and is more efficient when used for screening than small hairpin RNAs.
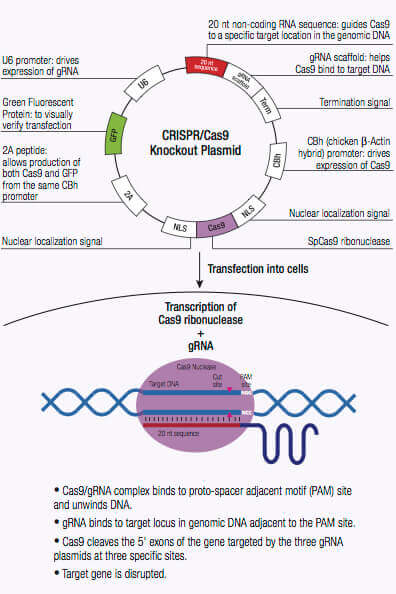
HDR plasmids provide a specific DNA repair template for a DSB, and are only used when co-transfected with CRISPR/Cas9 KO plasmids. When co-transfected with the CRISPR/ Cas9 KO plasmid, the HDR plasmid incorporates a puromycin resistance gene for selection of cells where Cas9-induced DNA cleavage has occurred.
The Cre Vector expresses Cre recombinase, a bacteriophage p1 enzyme that catalyzes site-specific DNA recombination between two LoxP sites. When the CRISPR/Cas9 knockout plasmid is co- transfected with the HDR plasmid, cells containing the edited DNA can be isolated using the selection marker inserted during homology-directed repair. Following selection, cells can be transfected with the Cre Vector to excise the genetic material inserted during homology-directed repair, such as the puromycin resistance gene.
These are the CRISPR/Cas9 knockout plasmids categories Santa Cruz Biotechnology offer:
We gladly support you by keeping you updated on our latest products and the developments around our services.